Uncovering Earth's Virome
Eric J. Ma (MIT)
Slides Online
IMED 2016
- 21st century disease emergence is accelerating, driven by population growth and related impact on the environment
- The world is still ill-prepared to respond to an emerging threat
- Our capacity to deploy effective “counter measures” is limited by what we “don’t know” about future threats
- Success will require changing from a culture that is “reactive” to one that is “proactive”
- We live in an age of Big Data which allows us to think differently about problems and solutions
- The Global Virome Project is about Big Data making a Big Impact
Motivating Questions
Metagenomics lets us answer these questions:
- How many viruses are out there?
- How can we discover them if we can't grow them?
Data Source and Processing
- Publicly available metagenomic data sets from the IMG/M1 system.
- 1,729 environmental samples
- 1,079 host-associated samples
- 234 engineered samples
Engineered: originating from the "built environment"; check "MicroBENet2".
Data Source and Processing
Problem:
- No universal markers, unlike barcode of life.
Solution:
- Leverage what we know:
- Protein families from 1,800 manually identified metagenomic viral contigs.
- Known viral protein families from isolates.
- Use known viral sequences as bait
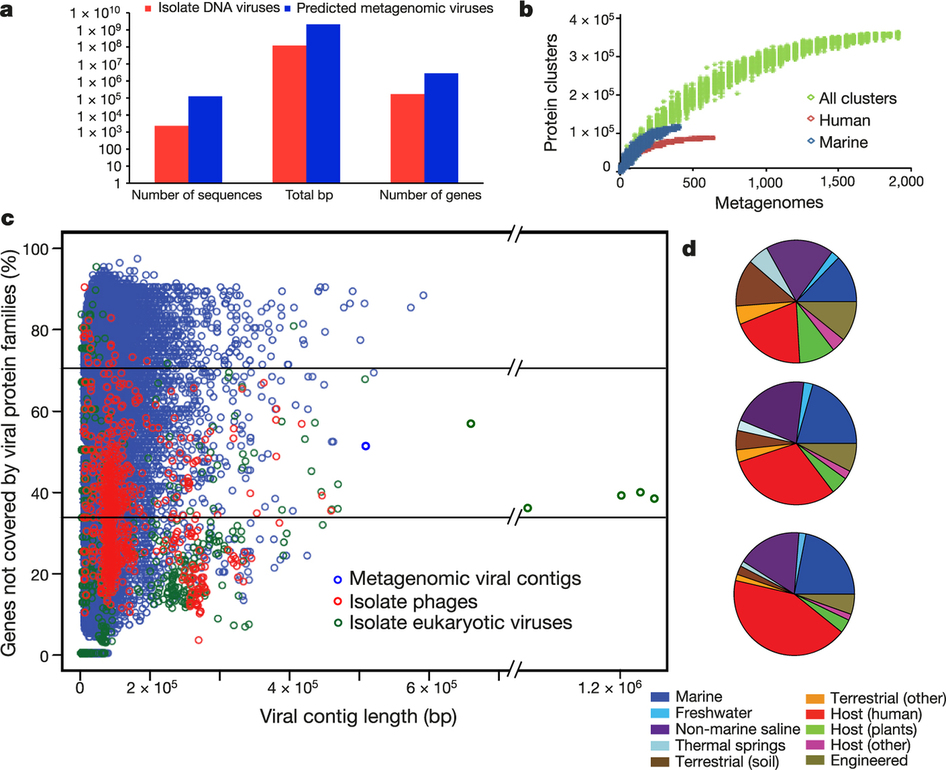
Metagenomic mining expands diversity of known viruses
- Large Viruses
- 596 kb contig, bioreactor
- Circular genomes by end overlaps
- 350-470 kb contig (6 samples)
Metagenomic mining expands diversity of known viruses
Host-virus interactions mapped by data integration
- Viral "Groups": Project known host-virus interactions.
- CRISPR-Cas: identify virus-prokaryote pairings.
- Viral tRNA: match to host genomes.
This is the one place where I think the paper uncovers new biology.
Expanded view of host-virus interactions
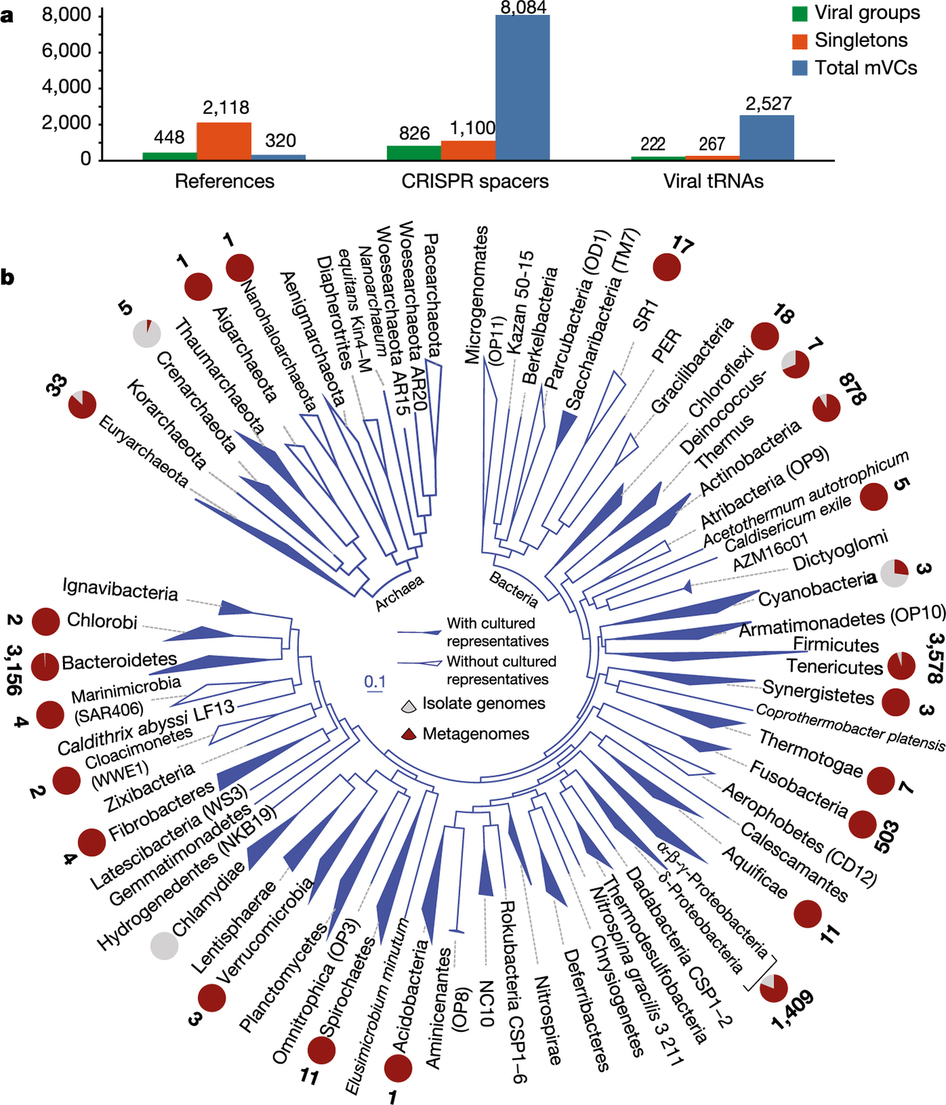
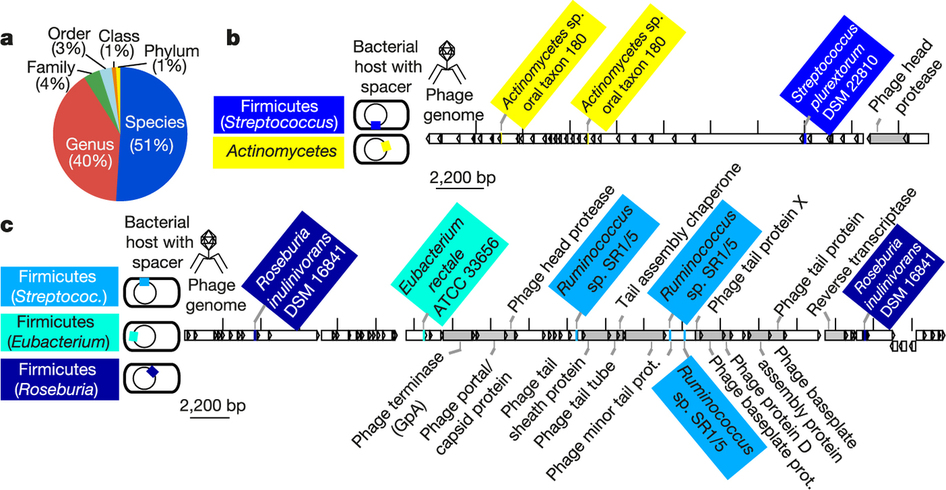
CRISPR spacers targeting same virus found on disparate phyla
Viral distribution is clustered according to habitat
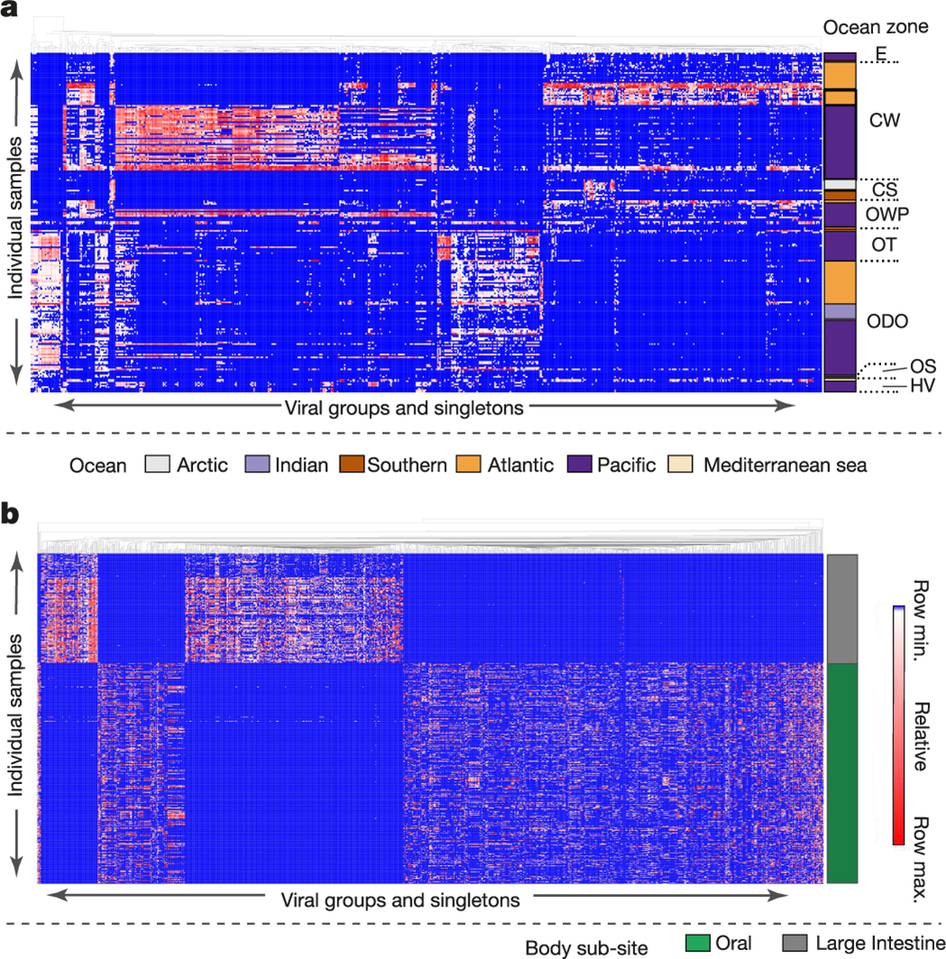
Viruses can be found basically all known environments
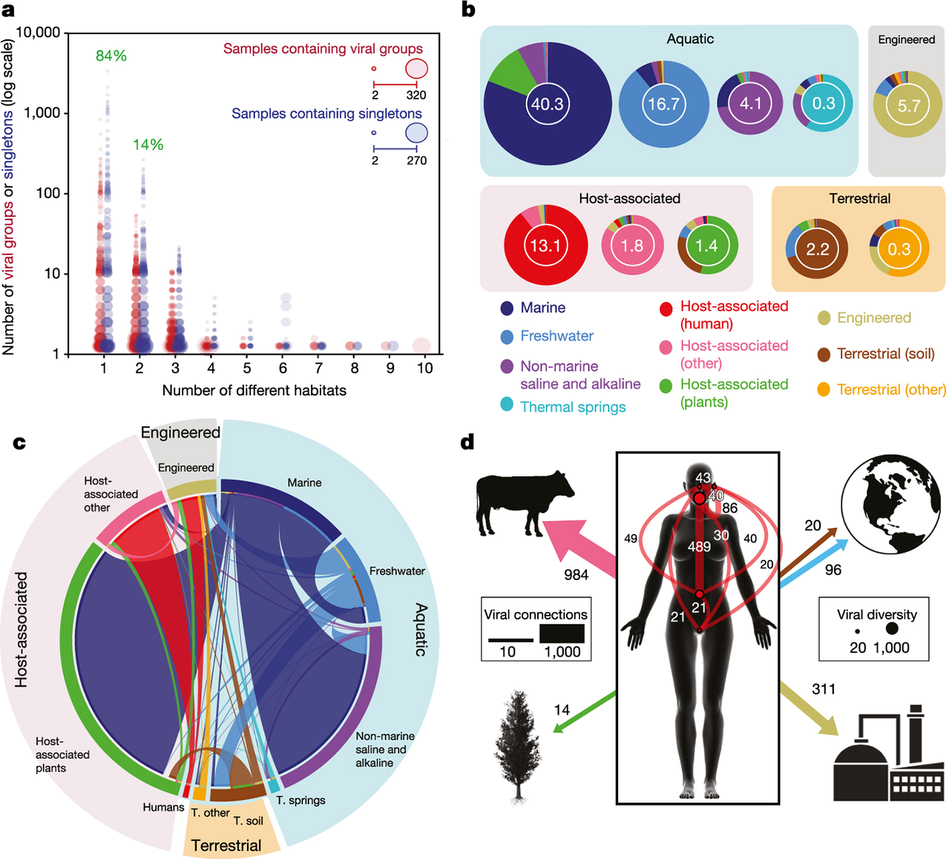
Viruses are shared between geographical locations
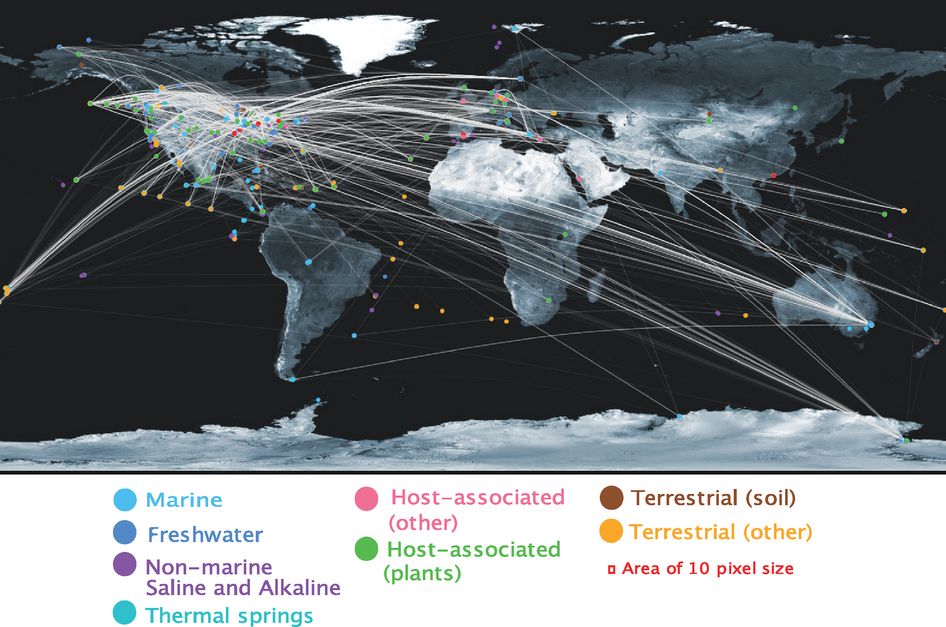
Paper Conclusions & Claims
- Metagenomics expands our view of the biosphere.
- Comprehensive map of host-virus interactions mined by combining datasets.
Personal Thoughts
On Influenza
- Influenza's host range is uniquely broad.
On Incentives
- Incentives for this study and its publication would not be provided by search for therapies.
- Yet therapies will very likely come out of this.