Eric J Ma's Website
written by Eric J. Ma on 2016-10-23 | tags: statistics data science
How we might make progress communicating statistics better.
There's the p-value vs. significance problem. I think it's widely agreed that education and training have a big role to play in fixing the culture of p-hacking. I think apart from the math-y parts of the problem, scientists are also stuck with a language problem.
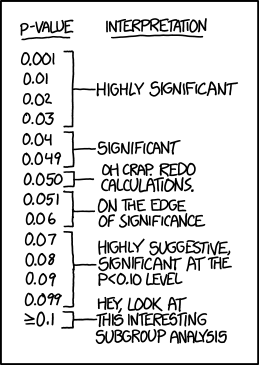
For example, I think you'd find this familiar: a statistically poorly-trained scientist will ask "whether the data show the hypothesis to be significant or not". I won't dive into an analysis as to how misleading the phrasing is, but I hope that the error in logical thinking is evident. Data don't decide whether a hypothesis is significant or not. The nature of the scientific question, and future historians' evaluation of the question, define the "significance" of the hypothesis being tested.
Less poorly informed scientists may ask, "is there a significant difference between my treatment and control?" That really depends on the magnitude of the difference, whether it's percentage/fractional/fold difference, or absolute difference, or odds ratios. Note here, p-values don't tell that. I'd point the reader to Allen Downey's excellent set of material that introduce this concept.
I'd assert here that for a scientist, the language that's used in describing the analyses and interpretations absolutely matter, and that taking away the "language of p-values" without providing something as a replacement will only result in the language of p-values returning. Here's my humble attempt at solving this problem, shamelessly borrowing ideas from other heroes in the PyData world.
(1) Instead of describing a "significant difference", describe the difference.
Replace the following phrasing:
our results showed a significant difference between treatment and control (INCORRECT)
with:
"the treatment exhibited 220% the activity level of the control". (CORRECT)
(2) Instead of reporting a p-value, report the effect size.
Following on from the statement, instead of saying:
our results showed that the treatment exhibited 220% the activity level of the control (p < 0.05) (INCORRECT)
replace it instead with:
our results showed that the treatment exhibited 220% the activity level of the control (cohen's d: 2.5) (CORRECT)
(3) Report uncertainty.
Let's build on that statement with uncertainty. Instead of:
our results showed that the treatment exhibited 220% the activity level of the control (cohen's d: 2.5) (INCOMPLETE)
replace it instead with:
our results showed that the treatment exhibited (220 ± 37%) the activity level of the control (100 ± 15%) (cohen's d: 2.5) (MORE COMPLETE)
(4) Go Bayesian.
With a generative model for the data (not necessarily a mechanistic one), uncertainty can be better represented in the figures and in the text. I'll show here just what the text would look like.
Here's the old version:
our results showed that the treatment exhibited (220 ±37%) the activity level of the control (100 ± 15%) (cohen's d: 2.5) (GOOD)
And here's the better version:
our results showed that the treatment exhibited 220% (95% HPD: (183, 257)) the activity level of the control (100%, 95% HPD: (85, 115)) (cohen's d: 2.5) (BETTER)
A quick thought on this final phrasing: The ± x% phrasing is inflexible for reporting distributions that may be more skewed than others, and assumes that the posterior distribution of the parameter is symmetrical.This is why I'd prefer reporting the 95% HPD, which if I'm not mistaken, is equivalent to the 95% confidence intervals when making certain prior assumptions in the Bayesian framework.
Update 24 October 2016: Just after making this post live, I saw a very helpful set of material by Justin Bois on how to decide which error bars to report. The ±x errorbars don't respect the bounds of the posterior distribution.
If accompanied by figures that report the estimated posterior distribution of values, we can properly visualize whether two posterior distributions overlap, touch, or are separated. That should provide the "statistical clue" about whether the results can differ from "statistical noise" or not. (Note: it still doesn't tell us whether the results are "significant" or not!)
What do you think about the need for language change?
Cite this blog post:
@article{
ericmjl-2016-reinventing-statistical-language-in-science,
author = {Eric J. Ma},
title = {Reinventing Statistical Language in Science},
year = {2016},
month = {10},
day = {23},
howpublished = {\url{https://ericmjl.github.io}},
journal = {Eric J. Ma's Blog},
url = {https://ericmjl.github.io/blog/2016/10/23/reinventing-statistical-language-in-science},
}
I send out a newsletter with tips and tools for data scientists. Come check it out at Substack.
If you would like to sponsor the coffee that goes into making my posts, please consider GitHub Sponsors!
Finally, I do free 30-minute GenAI strategy calls for teams that are looking to leverage GenAI for maximum impact. Consider booking a call on Calendly if you're interested!